Introduction to the Subject
Welcome to Environmental Informatics MATH3005 !
This book represents the lecture notes for this unit, if you're taking this class, you're expected to work your way through this book and also complete the corresponding Jupyter Notebooks.
Each Topic chapter corresponds to a topic in class and will list the required readings and any lecture notes to accompany the week.
Hypothesis Testing
The focus of this class should be a revision for all students.
There is no written material for this week, rather student's should refer to the allocated readings:
References
2 https://github.com/Chris-Engelhardt/data_sci_guide/blob/master/README.md
3 https://github.com/ossu/data-science#prerequisites
4 https://github.com/swirldev/swirl
Biased Estimators and Sample Statistics
Variance in Terms of Expected Differences
Variance is equal to the difference between the expected square value and the expected value squared:
Summing Variances
Note however, if and are not independent:
Multiplying Variance by a Constant}
Variance of a Sample Mean
You may recall this:
from the Central Limit Theorem 1, we develop this idea further in the section on t-distributions.
https://en.wikipedia.org/wiki/Central_limit_theorem
Consider a sample of size , the population will have a variance and a mean , the sample will have a variance and a mean . Many different samples could be taken, so there is a distribution of values that could occur. The variance of sample means is given by , this is a part of the CLT and is shown here.
Bessel's Correction
Suppose that the sample variance was given by the typical formula:
The expected value should be , in that sense it would be an un-biased estimator of the population mean.
The key here is to recognise that corresponds to a sample, by introducing we can solve for variance in terms of expected values and the sample mean. The sample introduces the bias so it is necessary to use the sample mean.
Note in particular, that is now a form of the expression, by the CLT this will give a , this is the key insight that pulls this together, it's that very that accounts for the :
Expected Value Squared
The expected value of is the mean value:
Expected Square Value
Recall the definition of Variance from earlier:
Applying this to the sample mean:
This step provides the key insight, if variance is taken on a population, there is no in the denominator
because the variance of a sample mean is different to the variance of a population statistic, the expected value of a squared sample mean is different to the expected value of a squared observation. The same from is the same one that leands to The expected value of a squared sample mean is less than the expected value of a squared observation, because the sample mean is going to be more central.
Solving The Expected Sample Variance
This shows that the variance formula, applied to a sample is biased. In order to correct that bias define like so:
The expected value of this is:
The t-distribution
Often is an unknown value, much like . It can be shown that replacing with results in a -distribution, so we often use that.
The t-distribution in terms of sample standard deviation
To show this, note the alternative form for :
Applying that to the typical standardization formula:
This right hant side is known as the t-distribution , this can be verified with R:
N = 3000
n = 5
qqplot(
rt(N, n - 1),
rnorm(N) / sqrt(n * rchisq(N, n - 1))
)
Sampling Distributions
Recall from Biased Estimators: Variance of a Sample Mean that:
Likewise:
Simple Linear Regression
There is no written material this week, rather see the readings:
References
1 Mendenhall, William, Robert J. Beaver, and Barbara M. Beaver. Introduction to Probability and Statistics. 14th ed./Student edition. Boston, MA, USA: Brooks/Cole, Cengage Learning, 2013.
2 https://www.openintro.org/book/os/
3 Murphy, Kevin P. Probabilistic Machine Learning: An Introduction. Adaptive Computation and Machine Learning Series. Cambridge, Massachusetts: The MIT Press, 2022.
4 Bingham, N. H., and John M. Fry. Regression: Linear Models in Statistics. Springer Undergraduate Mathematics Series. London ; New York: Springer, 2010.
Multiple Linear Regression
The content for this week consists primarily of readings. The subtopics included here show the derivation for Multiple Linear Regression.
Readings
References
1 https://www.openintro.org/book/os/
2 Mendenhall, William, Robert J. Beaver, and Barbara M. Beaver. Introduction to Probability and Statistics. 14th ed./Student edition. Boston, MA, USA: Brooks/Cole, Cengage Learning, 2013.
RSS as a Maximum Likelihood Estimator
If the residuals of a model are expected to be normally distributed, then the paramaters should be chosen to minimize the RSS 1.
Otherwise the choice of loss function is arbitrary.
Consider a linear regression:
If we presume that we could view this from a perspective of choosing a value that is normally distributed around with a variance of .
% If we treat as a a function of the probability of seeing a value , given that this model is true and the residuals are indeed normal, is given by , this would correspond to:
The probability of seeing any such value is:
The likelihood of seeing all the observations would be given by:
This function only has three parameters (, and ), everything else is either an observed value or a constant.
What we want to do is choose values of and that maximize this likelihood for any given :
Thus, in order to maximize the likelihood of seeing any with normally distributed residuals, it is sufficient to choose values and that minimize the RSS.
Footnotes
RSS: The Residual Sum of Squares
Matrix Chain Rule
If observations are recorded along rows (row-major), the following linear regression model holds and conventional definition of the partials:
For column-major the model is:
In either case, it will be shown that the chain rule will be:
Notation
The notation, is adapted from Knuth 1 and defined as:
Solving the Product Derivatives
So we have:
Solving the Chain Rule
For the column-major example :
This of course assumes column-major where the columns of represent observations and the rows are features, in the event that these were transposed we would have
and hence:
Graham, Ronald L., Donald Ervin Knuth, and Oren Patashnik. Concrete Mathematics: A Foundation for Computer Science. 2nd ed. Reading, Mass: Addison-Wesley, 1994.
Bias Terms in Multiple Regression
Consider a matrix multiplication as shown below.
Where:
- is the sample size
- is the number of columns in the output, i.e. the number of response variables (or dependent variables) being modelled
- is the number of columns in the input, i.e. the number of *features (or input/independent variables)
In this representation, each of the rows of and is an observation and the columns represent input and response variables respectively.
However, I have elected to use a column-major so let's instead represent this:
Now each observation is a column-vector instead of a row vector.
This corresponds to a matrix form thusly:
note the following points:
- This is a column-major representation, the vector represents
a single observation and subsequent observations would become additional
columns of .
- A row major representation can be acheived by transposing the matrices.
- R, Julia, Octave, Wolfram and Fortran all use column-major representations
- Python, C(++), Go and Rust use a row-major representation
- It's important to get this right, languages store values in a certain pattern in memory, cutting against the grain will be less performant.
- The use of sympy/octave/julia notation, whereby represents a vector composed of the first row of .
Let's now add a bias term (i.e. an intercept) so that the model reflects from simple linear regression:
The bias term could equally be expressed inside the matrix thusly:
That's why we don't often include a bias term when performing multiple linear regression, as it can be included as a component of the weights matrix.
More Observations
If there were observations:
Row Major
If we transformed this to a row-major representation, the 1s would now be a row of the input and the bias a row of the weights:
A note on Memory Layout in languages
Column major matrices are rooted in mathematical notation and conventions of linear algebra. In many textbooks and papers, 1D vectors are often treated as column vectors where convenient. This makes column-major order a natural and intuitive choice for mathematical languages where matrix operations are common. The reader may have already noted that many column-major languages are also 1-indexed, this is for a smiliar reason.
The C language was developed for systems programming, presumably row-major representation was chosen because it is consistent with how people usually layout data, there was no need to cater towards mathematical programming.
Deriving Multiple Linear Regression
Introduction
The goal here, is to maximize the probability of seeing some observation given a mean value and a constant variance
Deriving Multiple Linear Regression
Consider a multiple linear regression of the form:
The bias for this model is included in the weights matrix, as shown in Bias Terms in Multiple Linear Regression.
Consider the probability of seeing an observation under the assumption:
The goal in linear regression is to find such that:
The argument which maximises the likelhood of this expression is equivalent to the the argument which minimises the rss, see Maximum Likelihood is equal to Minimum RSS
where:
Note that the squaring operation is elment-wise, so more conventionally:
because is a quadratic function:
Recall the Matrix Chain Rule
Setting the derivative to 0:
Conclusion
So, in conclusion, if , the value for that minimises the l2 norm is 1 2 :
References
"5.4 - A Matrix Formulation of the Multiple Regression Model | STAT 462." Accessed August 17, 2023. https://online.stat.psu.edu/stat462/node/132/.
s 3.1 of Bingham, N. H., and John M. Fry. Regression: Linear Models in Statistics. Springer Undergraduate Mathematics Series. London ; New York: Springer, 2010.
Confidence and Prediction Intervals
- Ch. 3 Shmueli, Galit, and Kenneth C. Lichtendahl. Practical Time Series Forecasting with R: A Hands-on Guide. Second edition. Erscheinungsort nicht ermittelbar: Axelrod Schnall Publishers, 2018.
- 5.5 and 7.6 of https://otexts.com/fpp3/forecasting-regression.html 1
Also available in print form: Hyndman, Rob J., and George Athanasopoulos. Forecasting: Principles and Practice. Third print edition. Melbourne, Australia: Otexts, Online Open-Access Textbooks, 2021. https://otexts.com/fpp3/#.
Introduction
This lecture provides the reader with a general understanding of how to use statistical intervals as decision rules. It explains the different techniques used including confidence intervals, prediction intervals, tolerance intervals and control charts.
In environmental monitoring, these intervals are used to compare new observation values with a standard or background value. For example, suppose that a mechanical workshop applies to refinance the mortgage on the workshop, in such cases it is common to require a soil analysis for pollutants such as sulphuric acid 2. If the new measurements are greater than background values, is this evidence of an actual difference, or is it just noise?
To make an objective decision, we create a decision rule based on an interval. If the new values fall outside of the background interval, it suggests potential contamination. If the new values stay within the interval, there is insufficient evidence to suggest the higher measurements aren't caused by chance.
Footnotes
The electrolyte in lead-acid batteries used in modern cars (even in some electric cars to provide 12v to accessories)
Test Training Splits
Data Splits
Test Training Split
Cross-Sectional Data
When modelling data, we must use performance charactacteristics to fit a given model and we must also choose which model is most appropriate, consider the visualisation below 1:
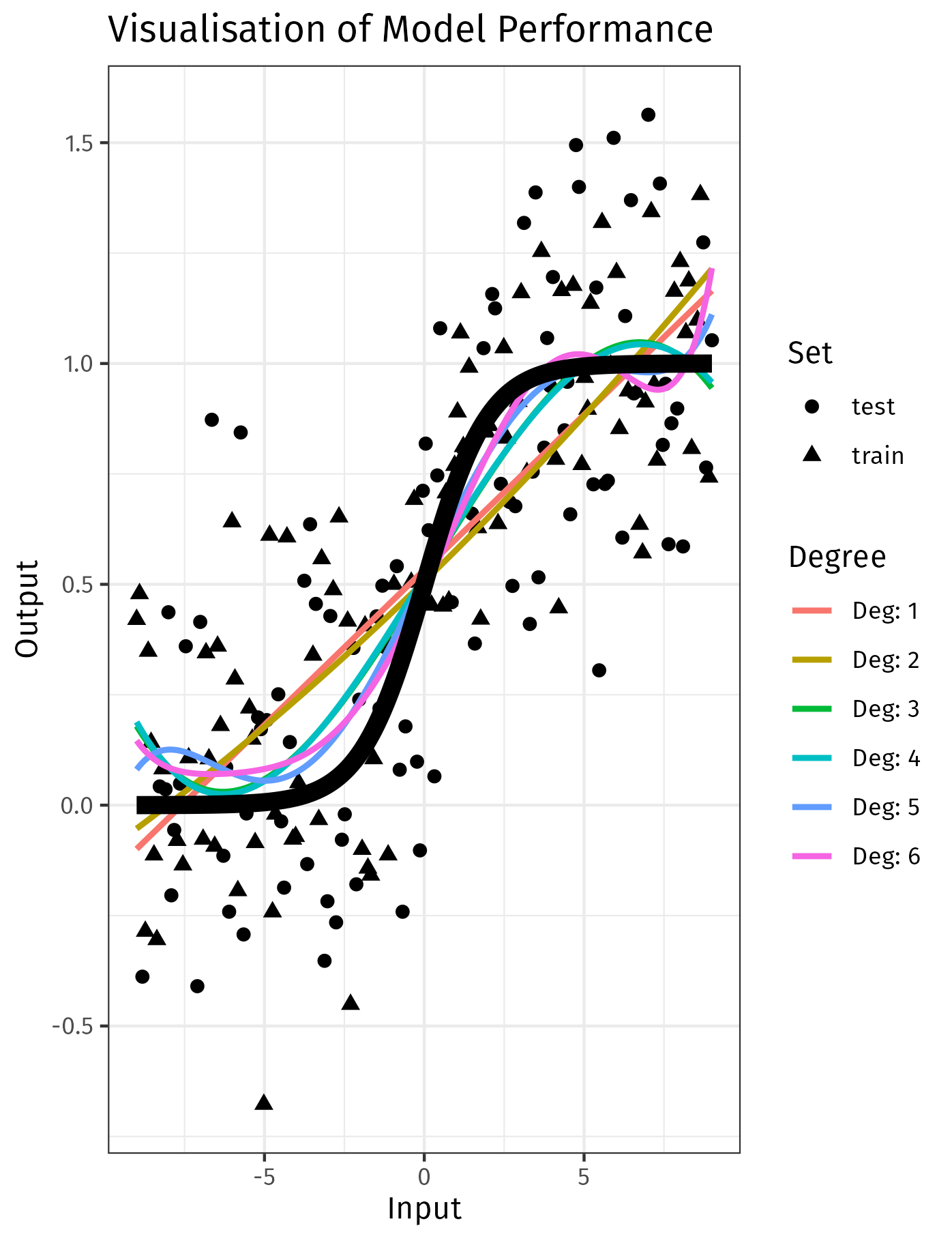
In that visualisation, models of a higher degree may fit the training data closer, but this occurs at the expense of general performance, i.e. validation error.
Typically we split data into three sets:
- Training
- Fit the model to this data
- Validation
- After fitting the model, measure the performance on this set of data
- This can be used to compare multiple models to determine which model was most accurate
- Testing
- After deciding on the best model, evaluate the performance on this data in order to estimate the amount of error in the model for unseen data.
There are other methods of estimating testing error AIC, BIC and cross validation, these aren't covered in this subject, see generally §§5.1, 6.1 of 2.
Cross-Sectional Data
Cross-Sectional Data
Firstly, set a seed for reproducibility, this can make debugging much easier 3:
set.seed(123)
data <- iris
Now, you need to create a random permutation of the row indices. This allows you to randomly shuffle the data:
index <- sample(1:nrow(data), nrow(data))
Use these indices to shuffle the dataset:
data <- data[index, ]
It's common to use 70% for training, 15% for validation, and 15% for testing. There is no hard rule for this though 4:
train_index <- round(0.7 * nrow(data))
valid_index <- train_index + round(0.15 * nrow(data))
Finally, use these indices to split the data:
train_data <- data[1:train_index, ]
valid_data <- data[(train_index+1):valid_index, ]
test_data <- data[(valid_index+1):nrow(data), ]
That's the general concept of testing and training splits. In practice it might be useful to use a dedicated library like caret, rsample or sklearn.model_selection.train_test_split in Python, these are less error prone because the logis is wrapped up in a well-tested function.
Also, be aware that splitting data this way might not maintain the original proportion of different classes especially in imbalanced type sets. For that case, you would need to do stratified sampling.
Also, be aware that this specific implemnetation did not maintain the orginial class proportions, this can be important for imabalanced datasets. When you have significant data imbalance, it may be necessary to look into other techniques such as over-sampling 5. An example of this type of situation is disease, models may simply predict all inputs as healthy when diseased observations are small in proportion.
Cross-Sectional Data
Split the data like so:
from sklearn import datasets
import sklearn
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import PolynomialFeatures
from sklearn.linear_model import LinearRegression
from sklearn.pipeline import make_pipeline
from sklearn.metrics import mean_squared_error
import numpy as np
# load iris datase
iris = datasets.load_iris()
# just use the first two features (sepal length and sepal width
X = iris.data[:, :2] # pyright: ignore
y = iris.target # pyright: ignore
# data spli
X_train_val, X_test, y_train_val, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
X_train, X_val, y_train, y_val = train_test_split(X_train_val, y_train_val, test_size=0.25, random_state=42)
We can visualize this test traiing split:
plt.scatter(X_train[:, 0], X_train[:, 1], c="r")
plt.scatter(X_val[:, 0], X_val[:, 1], c="b")
plt.scatter(X_test[:, 0], X_test[:, 1], c="g")
plt.legend(["Training", "Validation"])
plt.title("Train/Test/Val Split of Iris Data")
plt.xlabel(iris.feature_names[0])
plt.ylabel(iris.feature_names[0])
plt.savefig("../assets/images//test_training_split_iris_example.png")
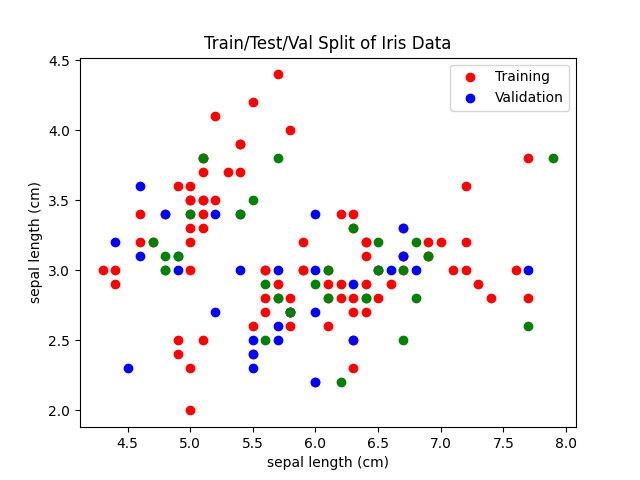
We can then fit a linear and polynomial model:
# Linear model
## Fit the model
model_linear = LinearRegression()
model_linear.fit(X_train, y_train)
## Get the Training Error
train_error_linear = mean_squared_error(
y_train,
model_linear.predict(X_train))
## Get the Validation Error
val_error_linear = mean_squared_error(
y_val,
model_linear.predict(X_val))
# Quadratic model
model_quadratic = make_pipeline(PolynomialFeatures(2), LinearRegression())
model_quadratic.fit(X_train, y_train)
## Get the Training Error
train_error_quadratic = mean_squared_error(
y_train,
model_quadratic.predict(X_train))
## Get the Validation Error
val_error_quadratic = mean_squared_error(
y_val,
model_quadratic.predict(X_val))
import json
my_dict = {
'training': {
'linear': round(train_error_linear, 3),
'quadratic': round(train_error_quadratic, 3) },
'validation': {
'linear': round(val_error_linear, 3),
'quadratic': round(val_error_quadratic, 3) }
}
print(json.dumps(my_dict, indent=2))
{
"training": {
"linear": 0.181,
"quadratic": 0.159
},
"validation": {
"linear": 0.209,
"quadratic": 0.202
}
}
Do you notice how the quadratic model has a lower training error (0.159 < 0.181) but the validation error is not signifianctly lower (0.209 ≅ 0.202)? This is a common pattern with complex models, they overfit the training data and perform pooly on validation data.
Temporal Data
Summary
When fitting models to time series data, it is also necessary to partition the data. However, in time series, the data is not split randomly, rather we allocate a portion of the most recent data as validation.
It is typical to omit a test period because the omission of the most recent periods may lead compromise the accuracy of the validation error, which may lead the wrong choice of model.
Example
Here is an example of a training/validation split applied to the EnvStats::Air.df data.
# Load required packages
library(xts)
library(EnvStats)
## Import some data as a dataframe
df <- EnvStats::Air.df
## First separate out the dates and the data
## use help(strftime) to find the format codes
index <- as.Date(rownames(df), format = "%m/%d/%Y")
data <- df
## converting df to xts
data_xts <- as.xts(df, order.by=index, format="%Y-%m-%d")
## Set split percentage
split_percent <- 0.75
## Get index to split the dataset
split_index <- round(nrow(data_xts) * split_percent)
## Create the training set
training_set_xts <- data_xts[1:split_index]
## Create the validation set
validation_set_xts <- data_xts[(split_index+1):nrow(data_xts)]
## Plot the Data
png(file = "../assets/images/validation_split_ozone_data.png",
width = 1024,
height = 1024,
units = "px")
plot.xts(na.approx(data_xts[ ,"wind"]))
split_event <- as.xts(
data.frame(
"Training-Validation Split",
order.by=as.Date(index[split_index])))
addEventLines(split_event, lwd = 5, col = "red")
dev.off()
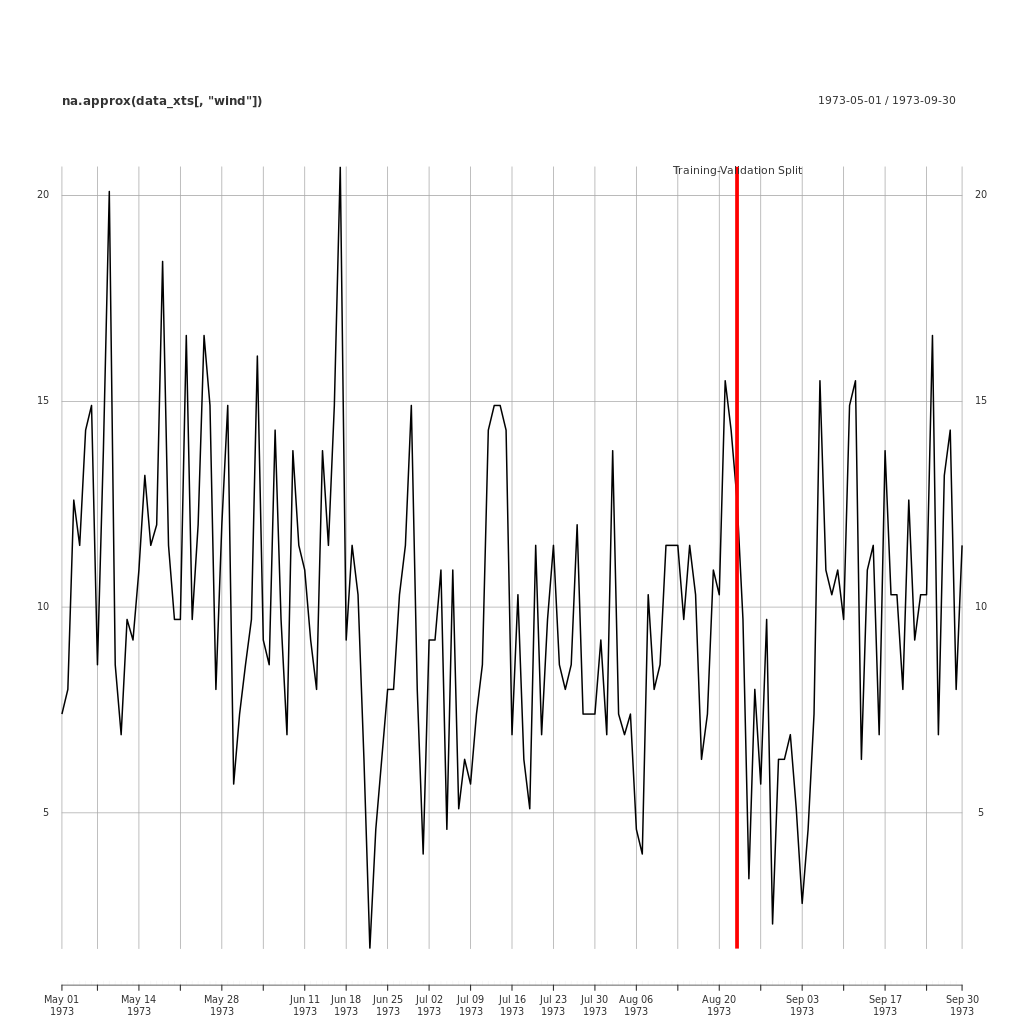
This code for this is included in the Appendix
Confidence Intervals
Summary
A Confidence Interval is a range of values, derived from a sample, within which the true population parameter is expected to lie. More precicely, the probability that a statistic taken from a random sample does not fall within a specified confidence interval is denoted by . In simpler terms, a confidence interval drawn from a sample, will capture the population statistic with a probability of . So in 20 samples, we would expect 19 of our confidence intervals to contain the population mean , iff the confidence intervals were 95%.
Take, for instance, a large yet finite population of fish. The average length of the fish in this population, while definite, is not possible to determine without measuring every single fish. If we were to draw 1000 samples, we'd expect that 900 of those sample means would fall within a 90% confidence interval.
It is important to be careful with the way we frame these probabilities. For example, if the data's 90% confidence interval ranged from 1 to 10, it would be incorrect to say that there's a 90% probability of the population mean falling within that range. This is because the population mean is a distinct, knowable value, it is not probalistic. The probability lies in the process of sampling. So, a more accurate statement would be, "if the population mean doesn't lie between 1 and 10, the probability of drawing such a sample was 10%".
The probability of a value within an interval, is given by the area under the curve for the corresponding distribution or PDF 1. Confidence intervals are no different, they simply correspond to a different distribution, the sampling distribution. Revisiting the fish population experiment, if we extract a sample of these fish and compute the sample mean, the obtained value will provide a good estimate for the population mean. If multiple samples are taken and their respective means are calculated, a distribution of these sample means can be developed. The area under the curve of this distribution corresponds to the probability of observing such a sample mean, this is interval is a confidence interval.
Probability Distribution
Derivation
The Central Limit Theorem states that if sample size (n) is large enough, typically n > 30 2, the distribution of sample means will approach a . Since the sample mean is known follows this distribution, the confidence interval represents a specific range within this distribution, capturing a predefined proportion of random events.
Often is an unknown value, much like . We've seen that replacing with results in a -distribution (see t-distribution), so confidence intervals will almost always be in terms of (unless the population variance is known, which is not common).
To derive the interval, re-arrange the z-score to represent bounds between an interval on the distribution:
Examples
Theoretical: Population Mean From Sample Mean
Consider a population, we can calculate the confidence interval using the t-scores:
## Construct a populatio
population <- c(
rep(0, 1000),
rep(2, 1000),
rep(30, 1000))
## Take a sampl
n = 30
samp = sample(population, n)
xbar = mean(samp)
s = sd(samp)
alpha = 0.05
t = abs(qt(alpha / 2, df=n-1))
# Compute the confidence limit
lower = xbar - t * (s / sqrt(n))
upper = xbar + t * (s / sqrt(n))
lower
upper
[1] 5.380967
[1] 15.81903
Note, the t.test function automatically performs a confidence interval:
## Conduct t-test and get confidence interval.
t_result = t.test(samp, conf.level = 0.95)
t_result$conf.int
If we repeat this many times, we can show that the confidence level is equal to the probability of an extreme sample:
## Construct a populatio
population <- c(
rep(0, 1000),
rep(2, 1000),
rep(30, 1000))
number_of_extreme_samples <- replicate(10^3, {
## Take a sample
n = 30
samp = sample(population, n)
xbar = mean(samp)
s = sd(samp)
alpha = 0.05
z = abs(qt(alpha / 2, df=n-1))
## Compute the confidence limits
lower = xbar - z * (s / sqrt(n))
upper = xbar + z * (s / sqrt(n))
lower
upper
lower < mean(population) && mean(population) < upper
})
mean(number_of_extreme_samples)
[1] 0.943
Bootstrapping: Confidence Intervals for Population Correlation
When a theoretical distribution for the sampling distribution is not known, it is possible to bootstrap a confidence interval. For example, consider the cars data:
| speed | list |
|---|---|
| 4 | 2 |
| 4 | 10 |
| 7 | 4 |
| 7 | 22 |
| 8 | 16 |
| 9 | 10 |
| ... | ... |
Let's assume that the R dataset is a population in and of itself. Let's take a sample use it to construct a confidence interval for the population mean. Unlike before, the formula for the confidence interval of correlation is not something most people have memorized. In this case we can bootstrap one.
The idea here is to assume that the population is an infinite repitition of the sample (acheived by sampling with replacement), we take samples from this bootstrapped population and use the quantiles of this psuedo-sampling-distribution as the confidence interval.
## Construct a populatio
population <- cars
head(cars)
n = 30
rho <- cor(population$speed, population$dist)
take_sample <- function(p) population[sample(seq_len(nrow(population)), n), ]
## Take a sampl
samp = take_sample(population)
rho_hat = cor(samp$speed, samp$dist)
## Write a function to produce a sampling distributio
make_sampling_dist <- function(samp) {
## Bootstrap a confidence Interval for that sample
replicate(10^3, {
## Take a sample with replacement (IMPORTANT)
boot_samp <- samp[sample(n, replace = TRUE), ]
## Calculate the sample statistic
cor(boot_samp$speed, boot_samp$dist)
})
}
get_conf_interval <- function(samp, alpha) {
lower = (1-alpha)/2
upper = 1-lower
quantile(make_sampling_dist(samp), c(lower, upper))
}
## Calculate the confidence interval
get_conf_interval(samp, 0.9)
5% 95%
0.6028544 0.8856464
Just like before, we can take this function for confidence intervals, and show that the probability of extreme samples is equal to the confidence of the interval:
## Construct a populatio
population <- cars
head(cars)
n = 30
rho <- cor(population$speed, population$dist)
take_sample <- function(p) population[sample(seq_len(nrow(population)), n), ]
## Take a sampl
samp = take_sample(population)
rho_hat = cor(samp$speed, samp$dist)
## Write a function to produce a sampling distributio
make_sampling_dist <- function(samp) {
## Bootstrap a confidence Interval for that sample
replicate(500, {
## Take a sample with replacement (IMPORTANT)
boot_samp <- samp[sample(n, replace = TRUE), ]
## Calculate the sample statistic
cor(boot_samp$speed, boot_samp$dist)
})
}
get_conf_interval <- function(samp, alpha) {
lower = (1-alpha)/2
upper = 1-lower
quantile(make_sampling_dist(samp), c(lower, upper))
}
## Calculate the confidence interval
get_conf_interval(samp, 0.9)
number_of_extreme_samples <- replicate(100, {
## Get a confidence Interval of a new sample
int <- get_conf_interval(take_sample(population), 0.8)
## Did that confidence Interval capture the population statistic?
int[1] < rho && rho < int[2]
})
mean(number_of_extreme_samples)
[1] 0.88
Unlike before, this value does not align with the confidence interval, that's because bootstrap methods are generally more likely to produce biased estimates when the sampling distribution is not normally distributed. This sampling distribution is right-skew, this could be inspected with:
hist(make_sampling_dist(samp))
There exist methods to overcome this limitation. 3
Linear Regression
Confidence Intervals of Linear Models
A linear model is premised on the assumption that the data follows a Gaussian distribution, characterized by a consistent variance, while the mean is a linear function as follows:
The difference with confidence intervals for distributions that we typically use is that here, the mean is a function. This interpretation also helps to better understand the underlying assumptions of linear regression. The constant variance and Gaussian residuals aren't so much assumptions of the model but rather integral characteristics of the model itself.
The confidence interval of a linear model, gives an interval through which we would expect different models to intersect. These models would correspond to different samples that we could have taken from the population.
Examples
Theoretical: Population Line From Sample Line
Consider the mtcars dataset:
head(mtcars)
mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1
A multiple linear regression can be fit and a confidence interval drawn for the parameters like so:
# Compute the confidence limit
f <- mpg ~ cyl + hp + wt
mod <- lm(f, data=mtcars)
# Get the confidence intervals for the linear model
confint(mod)
2.5 % 97.5 %
(Intercept) 35.0915623 42.412012412
cyl -2.0701179 0.186884238
hp -0.0423655 0.006289293
wt -4.6839740 -1.649972191
To predict a confidence interal on the values:
# Predict values and add confidence intervals
predict(mod, interval='confidence') |>
head()
fit lwr upr
Mazda RX4 22.82043 21.65069 23.99016
Mazda RX4 Wag 22.01285 20.98250 23.04319
Datsun 710 25.96040 24.71014 27.21066
Hornet 4 Drive 20.93608 19.94927 21.92289
Hornet Sportabout 17.16780 15.75195 18.58365
Valiant 20.25036 19.12109 21.37962
Bootstrap
Values
# Bootstrap the linear model and calculate the coefficient estimates
bootstrap_estimates <- replicate(10^3, {
## Sample the data with replacement
sample <- mtcars[sample(nrow(mtcars), replace = TRUE), ]
## Fit the linear model
mod <- lm(mpg ~ cyl + hp + wt, data = sample)
# Return the coefficients
predict(mod, newdata = list(cyl = 4, hp = 100, wt = 2.6))
})
alpha = 0.9
lower = (1-0.9)/2
upper = 1 - lower
quantile(bootstrap_estimates, c(lower, upper))
5% 95%
23.43200 26.48408
Parameters
This has been ommited, the focus here is contrasting with prediction intervals. To see an example of bootstrapping the parameters, refer to the index.
Derivation
Deriving confidence intervals for linear regression is the same concept as before but I won't be showing it here. See generally the first 4 chapters of 4.
Footnotes
PDF: Probability Density Function
It's worth noting that extremely non-symmetric distributions will require larger sample sizes. 30 is sufficent for symmetric data though.
If we wanted we could adjust it with Fisher's z-transformation which would help reduce the bias. This is shown in the Appendix: Fisher's z-tranform. More generally there is also bias-corrected and accelerated (BCa) bootstraps, this is a method to deal with bootstraps of any skewed sampling distribution.
Bingham, N. H., and John M. Fry. Regression: Linear Models in Statistics. Springer Undergraduate Mathematics Series. London ; New York: Springer, 2010.
Prediction Intervals
Summary
A prediction interval provides a range of values that a future individual observation will lie between, with a certain probability.
Prediction intervals are ranges that estimate the future individual observations to lie with a certain probability. These intervals are used in statistical modelling and machine learning to represent the uncertainty in predictions. This range differs from a confidence interval, which estimates the range of a population parameter rather than future observations.
Probability Distribution
Derivation
Examples
Normal Distribution
Problem
The list below shows aresenic concentrations (ppb) collected every two months at two groundwater monitoring wells:
| Well | Year | Arsenic (ppb) |
|---|---|---|
| Background | 1 | [12.6, 30.8, 52.0, 28.1] |
| Background | 2 | [33.3, 44.0, 3.0, 12.8] |
| Background | 3 | [58.1, 12.6, 17.6, 25.3] |
| Compliance | 4 | [48.0, 30.3, 42.5, 15.0] |
| Compliance | 5 | [47.6, 3.8, 2.6, 51.9] |
Summary statistics show year 4 has a higher sample mean and lower variance:
apply(df, 2, \(x) c("mean" = mean(x), "sd" = sd(x)))
| y1 | y2 | y3 | y4 | y5 | |
|---|---|---|---|---|---|
| mean | 30.87 | 23.27 | 28.40 | 33.95 | 26.47 |
| sd | 16.20 | 18.71 | 20.47 | 14.63 | 26.93 |
Is there evidence of contamination for the compliance well in year 4?
Theoretical
This section is adapted from 1
If is not known (which is likely), it can be shown 2:
This gives us:
This, however, is only a one step prediction. To take multiple predictions forward is yet more work. The 95% probability would correspond to the product of both observations and the interval would have to be adjusted accordingly. This is complicated to implement, so instead we use the Bonferroni Correction. Broadly speaking, the Bonferroni Correction offsets the fallacy of multiple comparisons by dividing the quantile by the number of comparisons, so instead the interval would be given by 3:
The help for EnvStats suggests this method will be more conservative:
Approximate Method Based on the Bonferroni Inequality (
method="Bonferroni")As shown above, when k=1k=1, the value of KK is given by Equation (8) or Equation (9) for two-sided or one-sided prediction intervals, respectively. When k>1k>1, a conservative way to construct a (1−α∗)100%(1−α∗)100% prediction interval for the next kk observations or averages is to use a Bonferroni correction (Miller, 1981a, p.8) and set α=α∗/kα=α∗/k in Equation (8) or (9) (Chew, 1968). This value of KK will be conservative in that the computed prediction intervals will be wider than the exact predictions intervals. Hahn (1969, 1970a) compared the exact values of KK with those based on the Bonferroni inequality for the case of m=1m=1 and found the approximation to be quite satisfactory except when nn is small, kk is large, and αα is large. For example, Gibbons (1987a) notes that for a 99% prediction interval (i.e., α=0.01α=0.01) for the next kk observations, if n>4n>4, the bias of KK is never greater than 1% no matter what the value of kk.
However, prediction intervals will always be too narrow 4, so using the Bonferroni correction is no less accurate and it is simpler to implement (even if libraries do it, we must verify our results). The EnvStats package includes both the bonferroni method and the exect method, for more information on the exact method see 5. The formula for the exact method is included below, but is not needed, this formula is adapted from 6:
Unnecessary math
Where:
- is the pdf of the standard normal distribution
- is the cumulative distribution function of the standard normal distribution.
Putting this all together, in R:
df <- data.frame(
y1 = c(12.6, 30.8, 52.0, 28.1),
y2 = c(33.3, 44.0, 3.0, 12.8),
y3 = c(58.1, 12.6, 17.6, 25.3),
y4 = c(48.0, 30.3, 42.5, 15.0),
y5 = c(47.6, 3.8, 2.6, 51.9))
y <- background <- c(df$y1, df$y2, df$y3)
compliance <- c(df$y3, df$y4)
pred_int_normal_population <- function(y, k, conf_level) {
n <- length(y)
a <- (1-conf_level) / 2
mean(y) + c(1, -1) * sd(y) * qt(p = a / k, df = n - 1) * sqrt(1 + 1/n)
}
pred_int_normal_population(y, k = 4, conf_level = 0.95)
## EnvStats::predIntNorm(x = y, n.mean = 1, k = 4, conf.level = 0.95)
[1] -25.54131 80.57465
Prediction Intervals of Other Models
Adapted from §5.5 of 7
where:
-
-
sd(y-yhat)
-
Different K values can be solved:
| Method | -step forecast standard deviation |
|---|---|
| Mean | |
| Naive | |
| Seasonal Naive | |
| Drift |
This will be handled for us by the forecast or EnvStats package.
Using predict
It's also possible to fit a stationary linear model, which is a mean-value time series, and call the predict function:
## y is the variable of concern
y <- rnorm(10)
## Generate some indices
x <- 1:10
## Fit a line
## The slope should be zero, so the model will fit only intercept
model <- lm(y ~ 0)
# Now create a new data frame with the new x values
new_x <- data.frame(x = 11:14)
# Use the 'predict' function to make prediction
predictions <- predict(model, new_x, interval = "prediction")
print(predictions)
EnvStats Package
df <- data.frame(
y1 = c(12.6, 30.8, 52.0, 28.1),
y2 = c(33.3, 44.0, 3.0, 12.8),
y3 = c(58.1, 12.6, 17.6, 25.3),
y4 = c(48.0, 30.3, 42.5, 15.0),
y5 = c(47.6, 3.8, 2.6, 51.9))
library(EnvStats)
EnvStats::predIntNorm(
c(df$y1, df$y2, df$y3), # Flatten the values
n.mean = 1, # 1 for 1 observation
k = 4, # Make 4 Future Predictions
conf.level = 0.95 # The probability for the interval
)
Results of Distribution Parameter Estimation
--------------------------------------------
Assumed Distribution: Normal
Estimated Parameter(s): mean = 27.51667
sd = 17.10119
Estimation Method: mvue
Data: c(df$y1, df$y2, df$y3)
Sample Size: 12
Prediction Interval Method: Bonferroni
Prediction Interval Type: two-sided
Confidence Level: 95%
Number of Future Observations: 4
Prediction Interval: LPL = -25.54131
UPL = 80.57465
Each of the values [48.0, 30.3, 42.5, 15.0] is within so there is insufficient evidence to conlcude there was contamination in year 4.
If we wanted to look over the next 8 years, the prediction interval would get wider:
library(EnvStats)
x = EnvStats::predIntNorm(
c(df$y1, df$y2, df$y3), # Flatten the values
n.mean = 1, # 1 for 1 observation
k = 8, # Make 4 Future Predictions
)
lpl <- x$interval$limits[1]
upl <- x$interval$limits[2]
obs <- c(df$y4, df$y5)
if (sum(lpl < obs & obs < upl)) {
print("No Evidence")
} else {
cat("Evidence to Reject H0")
}
Results of Distribution Parameter Estimation
--------------------------------------------
Assumed Distribution: Normal
Estimated Parameter(s): mean = 27.51667
sd = 17.10119
Estimation Method: mvue
Data: c(df$y1, df$y2, df$y3)
Sample Size: 12
Prediction Interval Method: Bonferroni
Prediction Interval Type: two-sided
Confidence Level: 95%
Number of Future Observations: 8
Prediction Interval: LPL = -32.47117
UPL = 87.50450
Log-Normal Distribution
All values are strictly positive, this may suggest a log-normal distribution is more appropriate.
Theoretical
This can be solved by using a log transform and then exponentiating the intervals:
pred_int_log_normal_population <- function(y, k, conf_level) {
n <- length(y)
a <- (1-conf_level) / 2
y <- log(y)
SE <- sd(y) * sqrt(1 + 1/n)
t <- qt(p = a / k, df = n - 1)
intervals <- mean(y) + c(1, -1) * t * SE
exp(intervals)
}
1.679715 278.144795
EnvStats
EnvStats::predIntLnorm(x = y, k = 4, conf.level = 0.95)
Results of Distribution Parameter Estimation
--------------------------------------------
Assumed Distribution: Lognormal
Estimated Parameter(s): meanlog = 3.0733829
sdlog = 0.8234277
Estimation Method: mvue
Data: y
Sample Size: 12
Prediction Interval Method: Bonferroni
Prediction Interval Type: two-sided
Confidence Level: 95%
Number of Future Observations: 4
Prediction Interval: LPL = 1.679715
UPL = 278.144795
>
Built-in Function: Population Value From Sample Value
Linear Regression
Prediction Intervals of Linear Models
Examples
Theoretical: Next Value from a Model
The prediction interval for a linear value is given by 8:
set.seed(123) # Ensuring reproducibility
n = 100 # Defining n
x = 1:n
m = 3
err = rnorm(n) # Creating error term
y = m * x + err # Generating y
data = data.frame(x = x, y = y) # Creating a data frame
x_i = 7 # This the point we are interested in
# Calculate prediction interval
fit = lm(y ~ x, data) # Fitting the model
predict(fit, newdata = list(x = x_i), interval = "predict", level = 0.95)
# Calculate prediction interval by hand
SE = summary(fit)$sigma # Extract the standard error from the model fit
hat_y_i = coef(fit)["(Intercept)"] + coef(fit)["x"] * x_i # Manual prediction using the model coefficients
t_val = qt(p = 0.975, df = n - 2) # t-value for 95% confidence level with n - 2 df [fn_df]
pred_int = hat_y_i + c(fit = 0, lwr = -1, upr = 1) * SE * sqrt(1 + 1/n + (x_i - mean(x))^2 / (n-1)*sd(x)) * t_val # Prediction Interval
# Results
pred_int
## footnotes
## [fn_df] Each parameter in the model consumes a degree of freedom
## 1 for intercept + 1 for slope
## See generally ch. 2 and 3 of
## Bingham, N. H., and John M. Fry. Regression: Linear Models in Statistics. Springer Undergraduate Mathematics Series. London ; New York: Springer, 2010.
fit lwr upr
20.98117 19.13687 22.82548
fit lwr upr
20.98117 19.13687 22.82548
In the interest of completeness, the SE could also be calculated from first principles:
## Define a function to get Standard Error from linear regression
linear_SE <- function(model) {
## Residual Sum of Squares (RSS)
## Could also do
## sum((y - predict(model))^2)
rss = sum(model$residuals^2)
# Degrees of Freedom
df = n - length(coef(model))
# Compute Standard Error (SE)
SE = sqrt(rss/df)
## Should be equal to
## Compare with SE from summary
## SE = summary(model)$sigma
}
Built-in Function: Next Value from a Model
Base Plots
# Build the linear mod
m <- lm(dist ~ speed, data = cars)
# Create a Prediction and Confidence interval for new data on the mod
alpha <- 0.95
newdata <- list(speed = cars$speed)
make_pred <- function(type) {
predictions = predict(m, newdata = newdata, interval = type, level = alpha)
data.frame(speed = cars$speed, predictions)
}
# Create datafra
pred_df <- make_pred("prediction")
conf_df <- make_pred("confidence")
# Plot the Da
plot(dist ~ speed,
data = cars,
xlim = range(speed),
ylim = range(dist),
main = "Prediction and Confidence Intervals",
xlab = "Speed",
ylab = "Distance"
)
# Add the prediction interv
cols = c(pred = "red", conf = "blue")
with(pred_df, {
lines(speed, fit, lwd = 2)
lines(speed, lwr, lty = 2, col = cols["pred"])
lines(speed, upr, lty = 2, col = cols["pred"])
})
# Add the Confidence Interv
with(conf_df, {
lines(speed, fit, lwd = 2)
lines(speed, lwr, lty = 2, col = cols["conf"])
lines(speed, upr, lty = 2, col = cols["conf"])
})
## Add a lege
legend("topright",
legend = c("Prediction", "Confidence"),
lty = c(2, 2), col = cols, lwd = 2
)
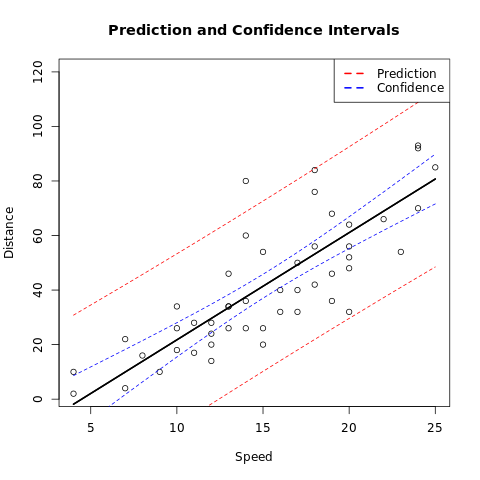
Tidyverse and GGPlot
# Load necessary librari
library(ggplot2)
library(tidyverse)
library(broom)
# Fit a linear model (Predict mpg using a single predictor, e.g., w
fit <- lm(mpg ~ wt, data=mtcars)
newdata <- data.frame(wt = seq(min(mtcars$wt), max(mtcars$wt), len=100))
conf_int <- predict(fit, newdata = newdata, interval = 'confidence') %>%
as_tibble() %>%
mutate(wt = newdata$wt)
pred_int <- predict(fit, newdata = newdata, interval = 'prediction') %>%
as_tibble() %>%
mutate(wt = newdata$wt)
# Making ggplot including confidence interv
ggplot() +
geom_point(data = mtcars, aes(x=wt, y=mpg)) +
geom_line(data=conf_int, aes(x=wt, y=fit)) +
geom_ribbon(data=pred_int, aes(x = wt, y = fit, ymin=lwr, ymax=upr), alpha=0.3, fill = "royalblue") +
geom_ribbon(data=conf_int, aes(x = wt, y = fit, ymin=lwr, ymax=upr), alpha=0.3, fill = "indianred") +
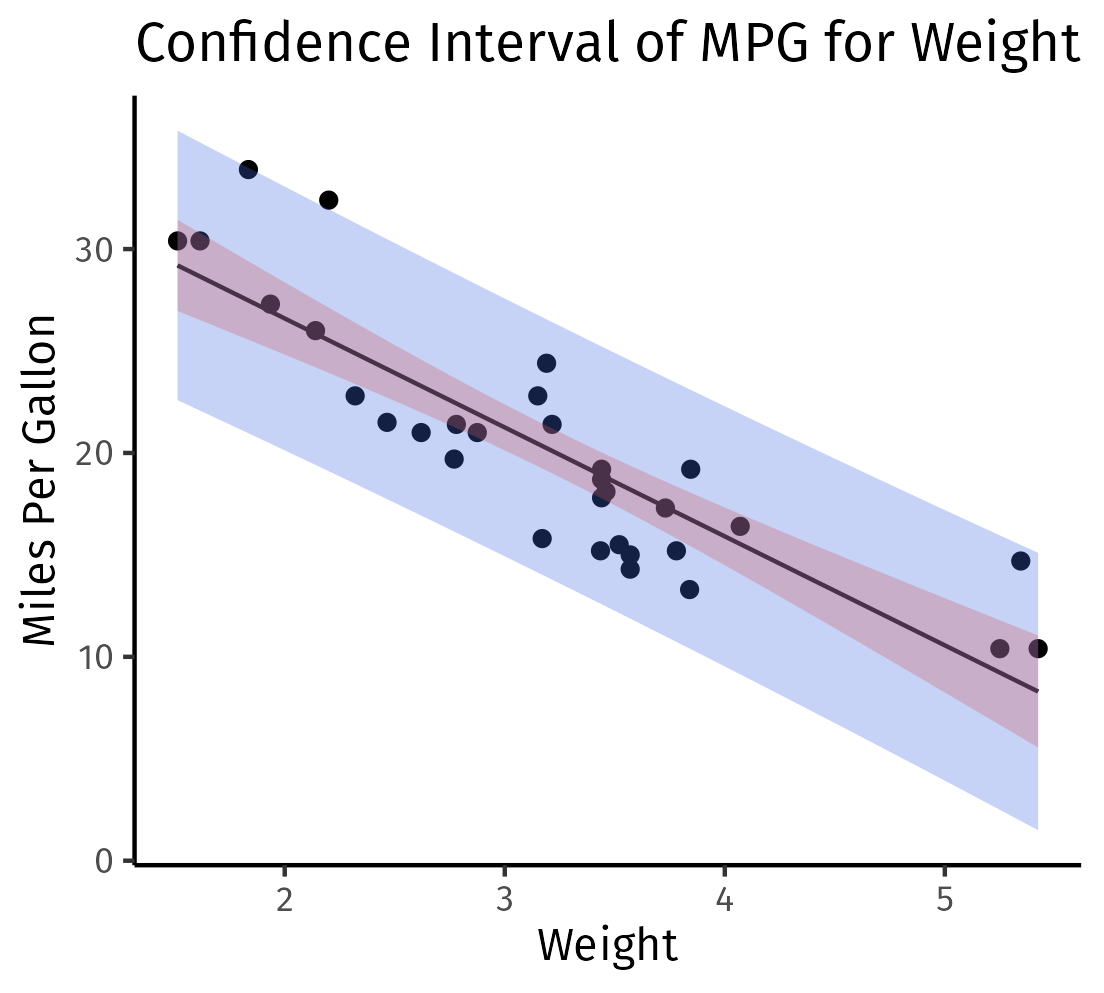
labs(title="Confidence Interval of MPG for Weight", x="Weight", y="Miles Per Gallon") +
theme_classic()
ggsave("../assets/images/confidence_and_prediction_interval.png")
Bootstrap
A simple bootstrap for the prediction interval of a linear model is included below. Note this assumes constant variance across the model, see 7 for a more detailed walkthrough of bootstrapping prediction intervals.
pred_vals <- replicate(10 ^ 4, {
n = nrow(my_cars)
form <- dist ~ speed
## Re-Sample the cars with replacement
s = sample(1:n, replace = TRUE)
cars_boot <- my_cars[s,]
## Fit a Linear Model
mod <- lm(form, cars_boot)
inter <- coef(mod)[1]
slope <- coef(mod)[2]
## Get the predicted value at x=10
pred_val <- 10 * slope + inter
## Find the residuals of the bootstrap model
residuals <- resid(mod)
## Randomly select one of the residuals
selected_resid <- sample(residuals, 1)
## Add the selected residual to the predicted value
pred_val_selected_resid <- pred_val + selected_resid
pred_val_selected_resid
})
hist(pred_vals)
quantile(pred_vals, c(0.025, 0.975))
## Confidence Interval
form <- dist ~ speed
fit <- lm(form, my_cars)
stats::predict(
fit,
newdata = list(speed = 10),
interval = "prediction",
levels = c(0.025, 0.975)
)
Derivation
Adapted from 9.
We define Linear regression for matrices and as:
The closed form solution for is:
We define:
- Population Slope
- Sample Slope
In the context of linear regression both and represent matrices of coefficients. If then the coefficient matrices will simply be vectors.
As a first step, we must solve the covariance of the weights matrix, before reading further make sure to review Covariance.
To begin, recall that the sum of variance in two vectors is the sum of the variance in each plus double the covariance:
We can decompose the weights matrix to include the population weights and the noise in the observation:
This can be used to show the weights matrix is an unbiased estimator of the population weights:
Re-arranging, this gives the difference between the sample and population weights matrix:
Finally, we can solve the covariance matrix in terms of the population standard deviation:
The confidence interval depends on the variance in model predictions:
This is the standard error used to solve the confidence interval.
The prediction interval is the standard error in the residuals, which is given by:
Using this we can solve the confidence interval with:
Of course, we don't know so we use t, it can be shown : 10
which implies
-
confidence interval:
-
prediction interval:
Where is the control matrix.
Footnotes
https://online.stat.psu.edu/stat501/lesson/3/3.3
https://en.wikipedia.org/wiki/Prediction_interval#Estimation_of_parameters
Dunnett, C.W. (1955). A Multiple Comparisons Procedure for Comparing Several Treatments with a Control. Journal of the American Statistical Association 50, 1096-1121. cited in 11
predIntNormK in https://search.r-project.org/CRAN/refmans/EnvStats/html/predIntNormK.html cited predIntNorm help page, see generally the manual Millard, Steven P. EnvStats: An R Package for Environmental Statistics. New York: Springer, 2013.
Rob J Hyndman's blog (Author of 7) https://robjhyndman.com/hyndsight/narrow-pi/ cited in https://joaquinamatrodrigo.github.io/skforecast/0.4.3/notebooks/prediction-intervals.html
Hyndman, Rob J., and George Athanasopoulos. Forecasting: Principles and Practice. Third print edition. Melbourne, Australia: Otexts, Online Open-Access Textbooks, 2021. https://otexts.com/fpp3/#.
Ch. 6, eq 6.22 Millard, Steven P., and Nagaraj K. Neerchal. Environmental Statistics with S-Plus. Applied Environmental Statistics. Boca Raton: CRC Press, 2001.
TODO, see generally https://en.wikipedia.org/wiki/Prediction_interval#Estimation_of_parameters
p. 89 § 3.7 of Bingham, N. H., and John M. Fry. Regression: Linear Models in Statistics. Springer Undergraduate Mathematics Series. London ; New York: Springer, 2010.
https://real-statistics.com/multiple-regression/confidence-and-prediction-intervals/confidence-and-prediction-intervals-proofs/
Tolerance Intervals
See 141 of EnvStats by Millard
Definition
A tolerance interval is a range that is likely to contain a specified proportion of the population, where ) is known as the coverage.
For example: Taking heights of students as an example.
Confidence Interval would be saying we are 95% confident that the population mean lies between 150cm and 200cm.
Tolerance Interval would be saying that we are 95% confident that 80% of the individual heights in the population lies between 150cm and 200cm.
The difference is that it isn't giving the range of values for a parameter estimate, rather it is a range where a proportion of future data points are likely to lie.
Constructing Tolerance Intervals
-content tolerance interval
This is constructed so that it contains at least of the poulation. i.e. the coverage is at least , with probability .
-expectation tolerance interval
This is constructed so that it contains on average of the population.
For a Normal Distribution
For normally distributed data, the upper () and lower () tolerance limits are computed form a series of measurements
Where is the tolerance factor:
- is the coverage ()
- is the t-distribution for , the significance level
- is the t-distribution for , the proportion coverage
Example in R
set.seed(222)
Y <- rnorm(100, mean=10, sd=2)
alpha <- 0.05 # significance level (95% CI)
P <- 0.85 # proportion of the population to be covered
n <- length(Y)
Ybar <- mean(Y)
s <- sd(Y)
## Calculating K tolerance factor
t_alpha <- qt(1-alpha/2, df = n-1)
t_P <- qt(1-(1-P)/2, df=n-1)
K <- t_alpha * sqrt(1 + 1/n + (t_P^2)/2/n)
U <- Ybar + K * s
L <- Ybar - K * s
print(paste("Tolerance Interval:", round(L, 4), round(U, 4)))
[1] "Tolerance Interval: 6.2033 13.8846"
Compare this with the EnvStats package
library(EnvStats)
tolIntNorm(Y, coverage=P, conf.level=1-alpha)$interval$limits
LTL UTL
6.900067 13.187851
They are close but not equivalent, because the EnvStats package uses a slight different, more complicated algorithm (See 1).
But from this, we can see that with 95% confidence, 85% of the population will lie between 6.9 and 13.2
https://rdrr.io/cran/EnvStats/src/R/tolIntNormK.R
Control Charts
Control charts are a graphical and statistical method of assessing the performance of a system over time.
They were developed in the 1920s by Walter Shewhart and have been employed widely in industry to maintain process control.
However, control charts assume the observations are independent and follow a normal distribution with some constant mean and standard deviation
Shewhart Control Chart
A Shewhart control chart is to plot the observations over time and compare them to established upper and/or lower control limits that are based on historical data.
Once a single observation falls outside the control limit(s), this is an indication that the process is "out of control" and needs to be investigated.
The constant is often set to , and the limits are called 3-sigma control limits
CUSUM Charts
To detect a gradual trend in the process, we may use Cumulative Summation (CUSUM) charts.
For the future sampling occasion, the upper cumulative sum and lower cumulative sum
Where is a given positive threshold that corresponds to half the size of a linear trend (in units of standar deviations), dependent on how sensitive to detection.
With a CUSUM chart, we declare a process "out of control" when the upper/lower cumulative sums are more extreme that a pre-specified decision bound, called the decision interval. Typically this is between 4 and 5.
Example in R
## Writing out the data
month <- 1:8
## baseline values in 1995
baseline <- c(32.8, 15.2, 13.5, 39.6, 37.1, 10.4, 31.9, 20.6)
## compliance values in 1996
compliance <- c(19, 34.5, 17.8, 23.6, 34.8, 28.8, 43.7, 81.8)
nickel <- data.frame(month, baseline, compliance)
month baseline compliance
1 1 32.8 19.0
2 2 15.2 34.5
3 3 13.5 17.8
4 4 39.6 23.6
5 5 37.1 34.8
6 6 10.4 28.8
7 7 31.9 43.7
8 8 20.6 81.8
## summary estimates from baseline
mean(baseline)
sd(baseline)
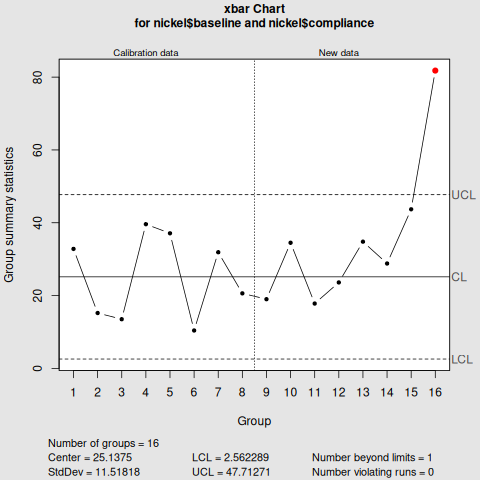
Shewhart Control Chart
library(qcc)
qcc(nickel$baseline,
type="xbar",
std.dev=sd(nickel$baseline),
newdata=nickel$compliance,
nsigmas=3,
confidence.level=0.95)
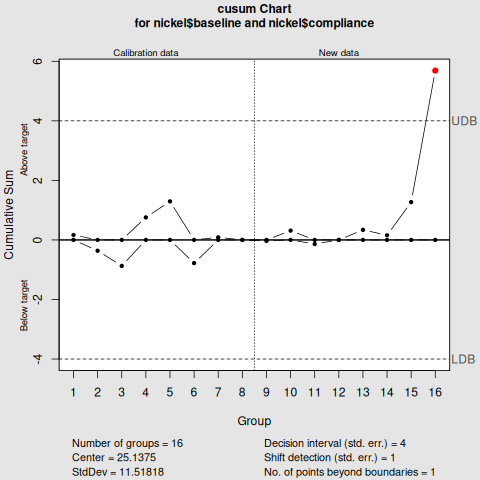
CUSUM Control Chart
cusum(nickel$baseline,
std.dev=sd(nickel$baseline),
decision.interval=4,
se.shift=1,
newdata=nickel$compliance)
Interpretation
Notice that the key difference between the two is the nsigmas in Shewhart control chart, and the decision.interval in the cusum control chart.
From the two visualisations, month 8 exceed the upper limit, deeming the nickel concentration "out of control".
Summary
- Confidence Interval gives an estimate of a population parameter
- Prediction Interval predicts where a single future observation will fall
- Tolerance Interval gives a cover of where a specified proportion of the poulation will fall.
- Control Charts have been suggested as an alternative to prediction or tolerance intervals, for determining whether a process is in a state of statistical control.
Displaying Dates
Displaying Dates
We haven't discussed time series yet, when do, we will need to describe dates. There are countless ways to format a date but there is only one way we will do it, ISO8601 1. In the ISO8601 we represent dates like so:
2006-01-02T15:04:05Z07:00
This corresponds to:
Mon Jan 2 15:04:05 MST 2006
Displaying Dates
strftime
The more common way to format time is strftime. This approach uses a collection of symbols to represent components of time, for a list of these symbols see man strftime 2, R's ?strftime and Python's help(time.strftime):
man strftime
The characters of ordinary character sequences (including the null byte) are copied verbatim from format to s. However, the characters of conversion specifications are replaced as shown in
the list below. In this list, the field(s) employed from the tm structure are also shown.
`%a` The abbreviated name of the day of the week according to the current locale. (Calculated from tm_wday.) (The specific names used in the current locale can be obtained by calling
nl_langinfo(3) with ABDAY_{1–7} as an argument.)
`%A` The full name of the day of the week according to the current locale. (Calculated from tm_wday.) (The specific names used in the current locale can be obtained by calling nl_lang‐
info(3) with DAY_{1–7} as an argument.)
`%b` The abbreviated month name according to the current locale. (Calculated from tm_mon.) (The specific names used in the current locale can be obtained by calling nl_langinfo(3) with
ABMON_{1–12} as an argument.)
`%B` The full month name according to the current locale. (Calculated from tm_mon.) (The specific names used in the current locale can be obtained by calling nl_langinfo(3) with
MON_{1–12} as an argument.)
The strftime approach was named after the C library that popularized it, it's used in C++, Rust, R, Python, PHP, Ruby and many more.
To represent in ISO8601 via strftime, the format would be %Y-%m-%dT%H:%M:%S, so e.g.:
-
R
format(Sys.time(), "%Y-%m-%dT%H:%M:%S") -
Python
from datetime import datetime now = datetime.now() formatted = now.strftime("%Y-%m-%dT%H:%M:%S") print(formatted) -
Shell
# ISO8601 date "+%Y-%m-%dT%H:%M:%S" # Unix date %+s
More Languages
-
Rust
extern crate chrono; use chrono::offset::Local; use chrono::DateTime; fn main() { let now: DateTime<Local> = Local::now(); println!("{}", now.format("%Y-%m-%dT%H:%M:%S").to_string()); } -
Julia
using Dates println(Dates.format(now(), "yyyy-mm-ddTHH:MM:SS")) -
C
#include <stdio.h> #include <time.h> int main() { char buffer[80]; time_t t = time(NULL); struct tm *tm_info; tm_info = localtime(&t); strftime(buffer, 80, "%Y-%m-%dT%H:%M:%S", tm_info); printf("%s\n", buffer); return 0; }
You'll notice that each language (except Go and Julia) uses a similar approach to format the time, standardized by the strftime approach.
Reference Time
In this approach the language requires a specific date to format time, the specific date (Mon Jan 2 15:04:05 MST 2006) was a design decision made by the development team because they are distinct in the context of a date -- they used each number from 0 to 6 only once:
2006: Represents the full year format.01: Stands for the numerical month format.02: Represents the day of the month.15: Stands for the 24-hour format time.04: Represents the minutes.05: Represents the seconds.MST: Time zoneMonandJan: Are used for the abbreviated day of the week and month respectively.
Reference time is not a common approach, it's more of a unique quirk of the Go language. We draw attention to it here because it is a convenient way to express time in a human readable way, this can be useful when looking up documentaion, researching or querying a LLM. For example, to print the current time in ISO8601 format we would write it like this:
package main
import (
"fmt"
"time"
)
func main() {
t := time.Now()
fmt.Println(t.Format("2006-01-02T15:04:05-0700"))
}
Naive Forecasts
Measuring Prediction Accuracy
Roll Forward Validation
Time Series Analysis
- Ch. 3-5 of https://otexts.com/fpp3/decomposition.html
- Ch. 2-5.3 of Shmueli, Galit, and Kenneth C. Lichtendahl. Practical Time Series Forecasting with R: A Hands-on Guide. Second edition. Erscheinungsort nicht ermittelbar: Axelrod Schnall Publishers, 2018.
- 3-5 of Rob J., and George Athanasopoulos. Forecasting: Principles and Practice. Third print edition. Melbourne, Australia: Otexts, Online Open-Access Textbooks, 2021. https://otexts.com/fpp3/#.
- ss 1.2, 1.8, 1.11, 1.3, 1.4, 1.5, 1.14, 1.15 of tsa3
-
Visualisation
-
Time Series Decomposition
- Components (Trend, Cycle, Season)
- Classical
- Additive
- Multiplicative
- Decomposition by Statistical Agencies, we won't cover these
- X-11
- SEATS
- STL Decomposition
-
Autocorrelation
- ACF
- PACF
-
Functions in Time Series
- Mean and Variance
- Covariance
- Auto-Covariance
- Autocorrelation
-
Processes
- Stationary Time Series
- White Noise
- Random Walk
- Drift
- Moving Average
- Auto Regressive
-
Simple Forecasting Methods
- Naive Method
- Mean Method
- Residual Diagnostics
google_2015 |> model(NAIVE(Close)) |> gg_tsresiduals()
-
Checklist:
-
Merge old notes
06.docx - Merge Stoffer's TSA3
-
Merge Lecture Slides
SP2020.pptx-
Start with
~/Studies/Teaching/2023/Spring/Env_Info/environmental-informatics_private/New_Material/06/todo/6_Time Series and Autocorrelation (Wk 6)/Lecture 6.docx, much of the slides are built off that.
-
Start with
- Merge fpp3
-
Merge old notes
Data Driven: Smoothing Methods
Moving Average
Exponential Smoothing
State Space Models
Model Based: ARIMA Models
Appendix
Appendix
Multiple overlayed models
library(tidyverse)
## Make some data.........................................
n <- 200
x <- seq(from = -9, to = 9, length.out = n)
y_t <- exp(x) / (1 + exp(x))
y <- y_t + rnorm(n, mean = 0, sd = 0.3)
i <- sample(c(TRUE, FALSE), size = n, replace = TRUE)
data <- tibble(x, y, Set = c("train", "test")[i %% 2 + 1])
## Construct some polynomial models.......................
preds <- list()
for (i in 1:6) {
## [glue]
mod <- lm(y ~ poly(x, i), filter(data, Set == "train"))
data <- data %>%
mutate("Deg: {i}" := predict(mod, newdata = data["x"]))
}
## Plot the data..........................................
data_tidy <- data %>%
select(!c(y, Set)) %>%
pivot_longer(cols = !x, names_to = "Degree")
data_tidy
ggplot(data_tidy) +
geom_point(data = data, aes(x = x, y = y, shape = Set), size = 2) +
geom_line(aes(x = x, y = value, color = Degree), lwd = 1) +
geom_line(data = tibble(x, y = y_t), aes(x = x, y = y), lwd = 3) +
theme_bw() +
labs(x = "Input", y = "Output",
title = "Visualisation of Model Performance")
ggsave("many_model_plot.png")
## [glue] This is glue like syntax available in dplyr>=1.
Fisher's Z Transform
get_conf_interval <- function(samp, alpha) {
lower = (1-alpha)/2
upper = 1-lower
z <- make_sampling_dist(samp)
## z is not normally distributed, we could improve accuracy with
## Fisher's z-transformation
z <- 0.5 * log((1 + z) / (1 - z))
quantile(z, c(lower, upper))
}